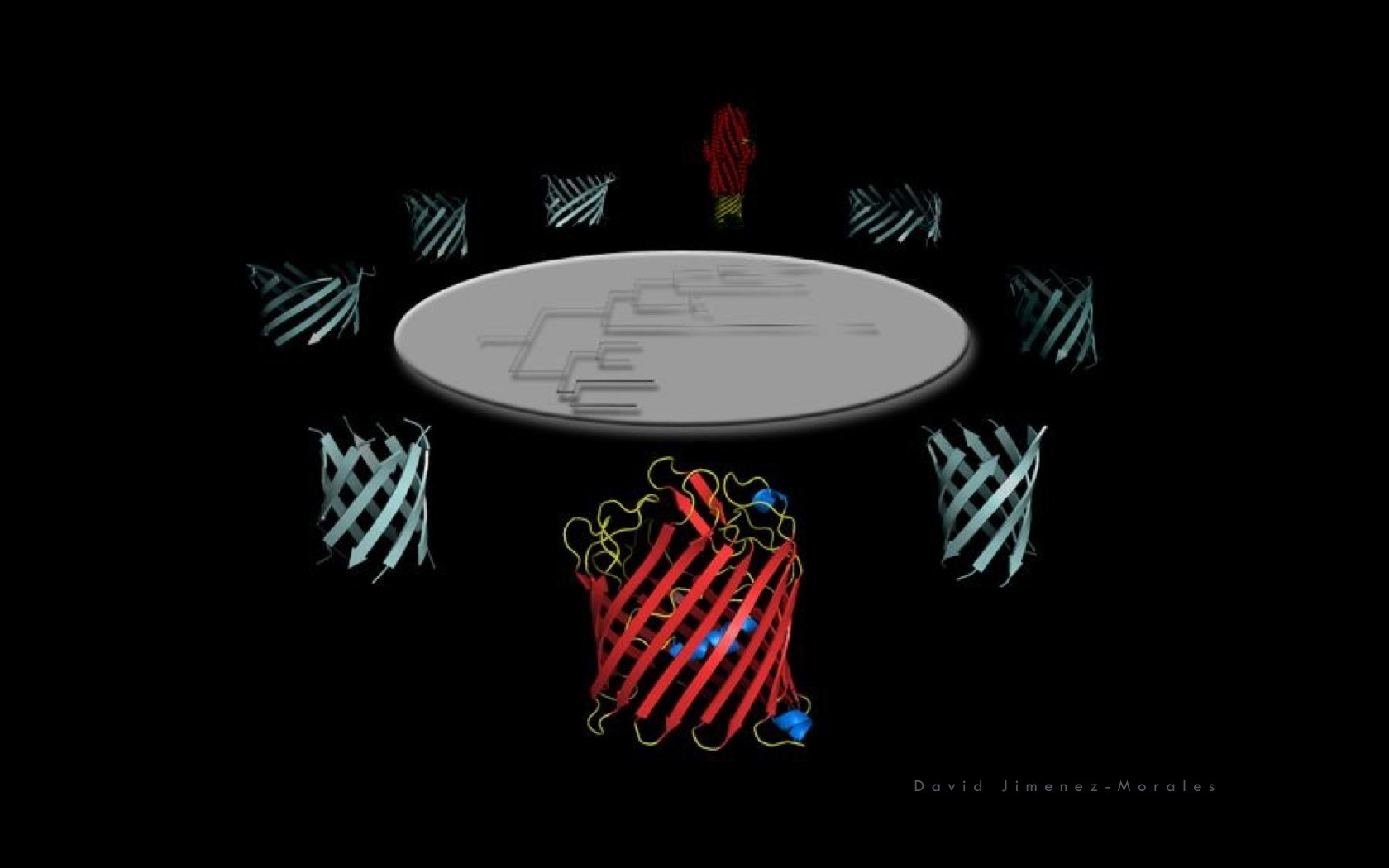
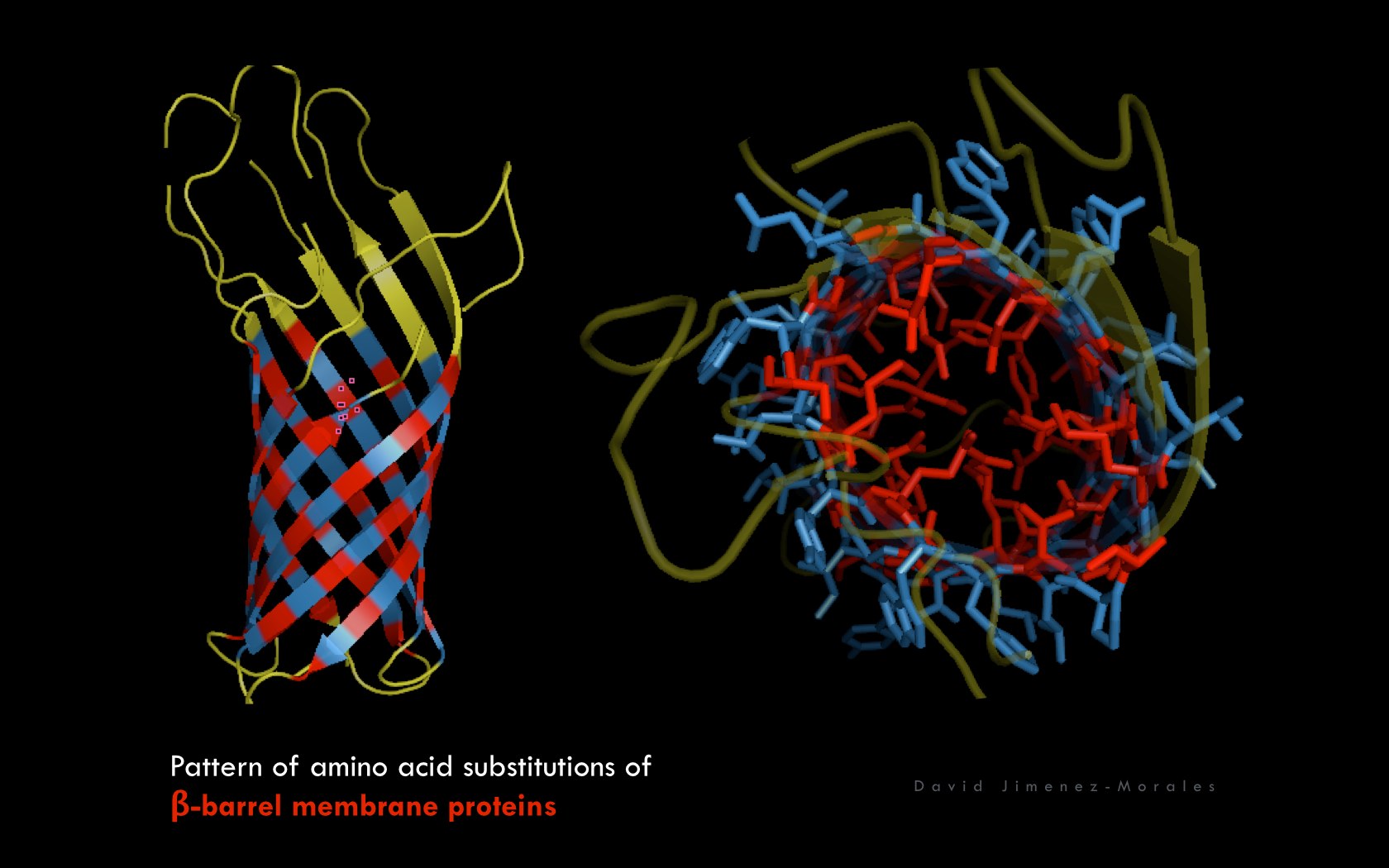
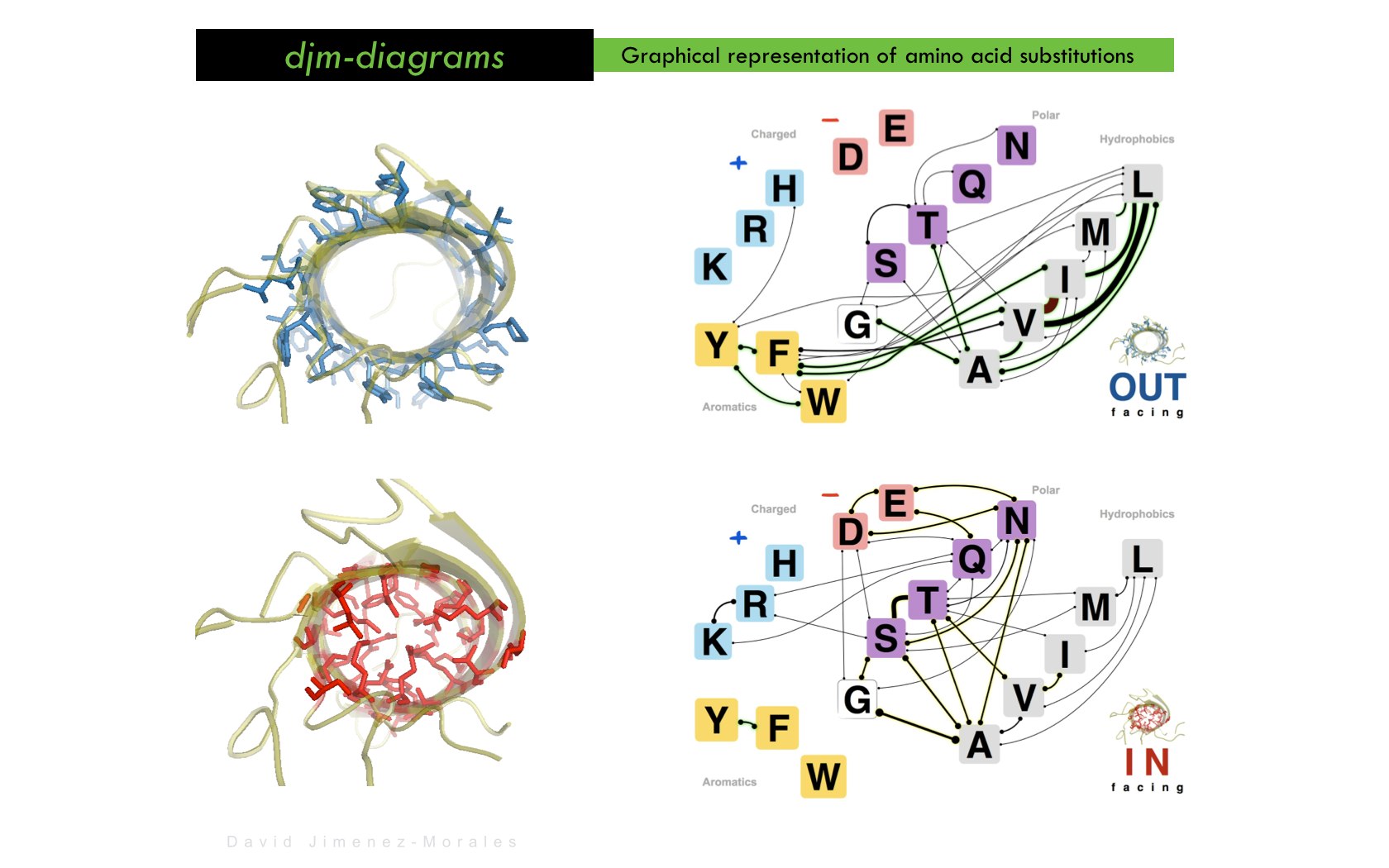
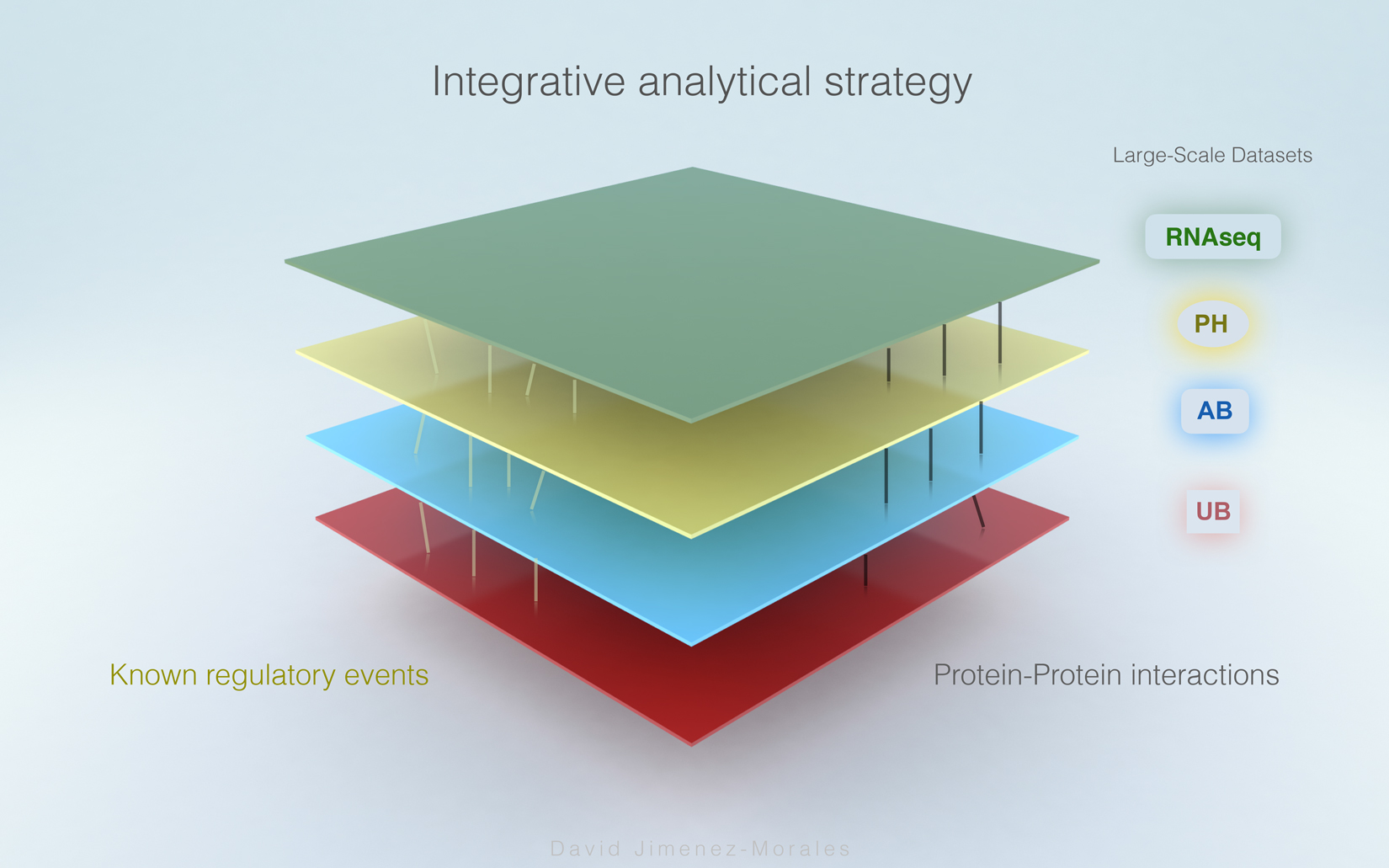
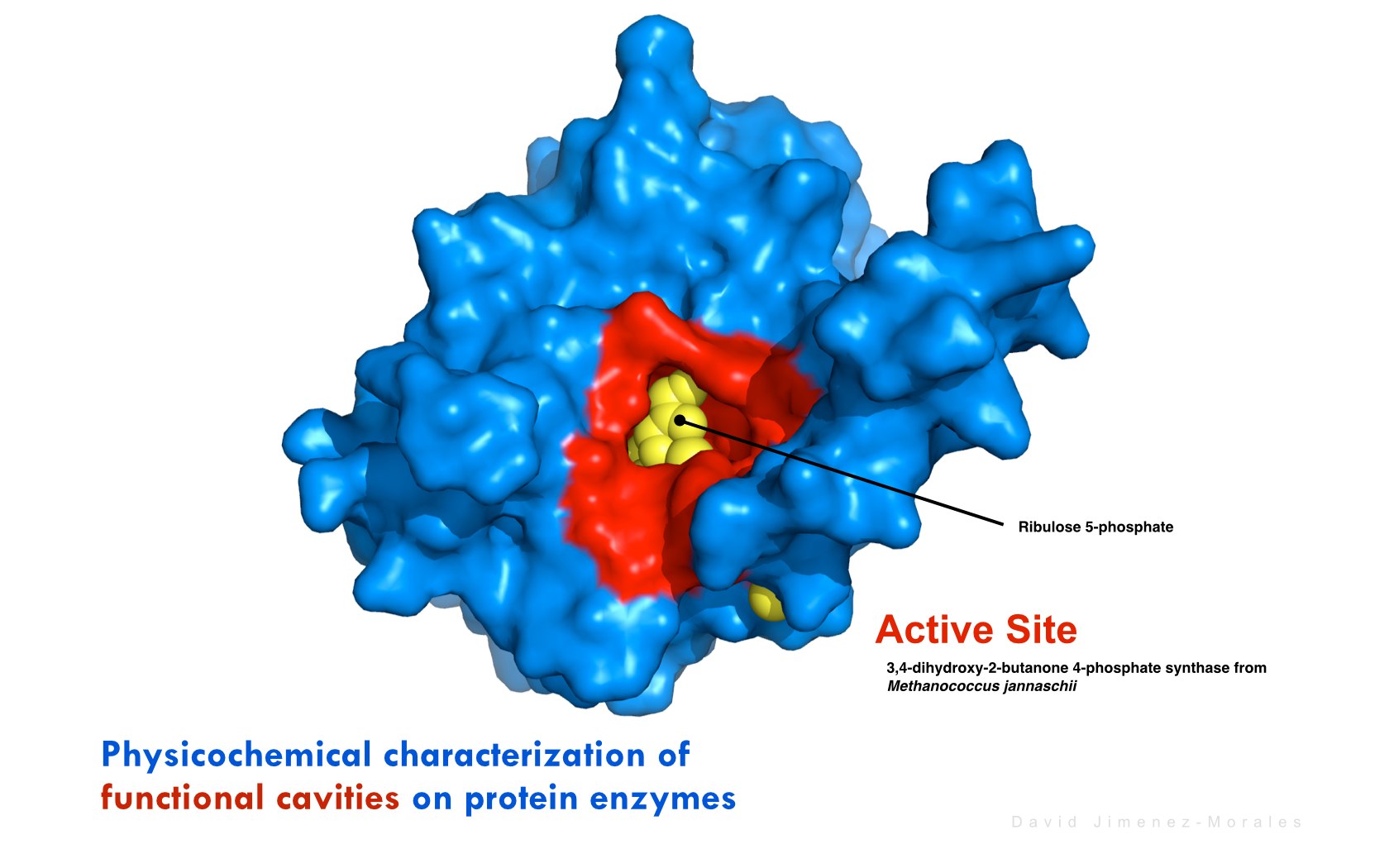
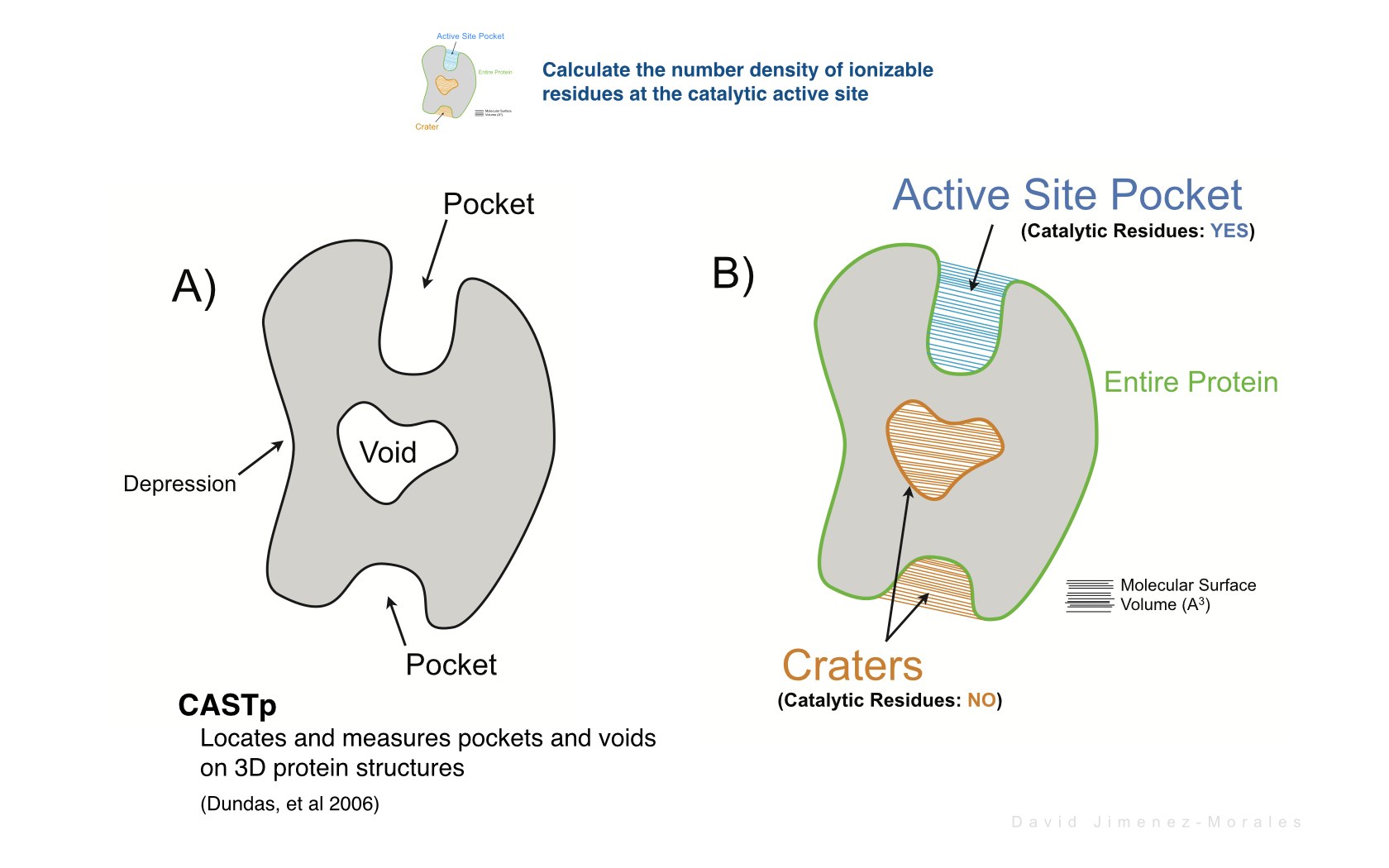
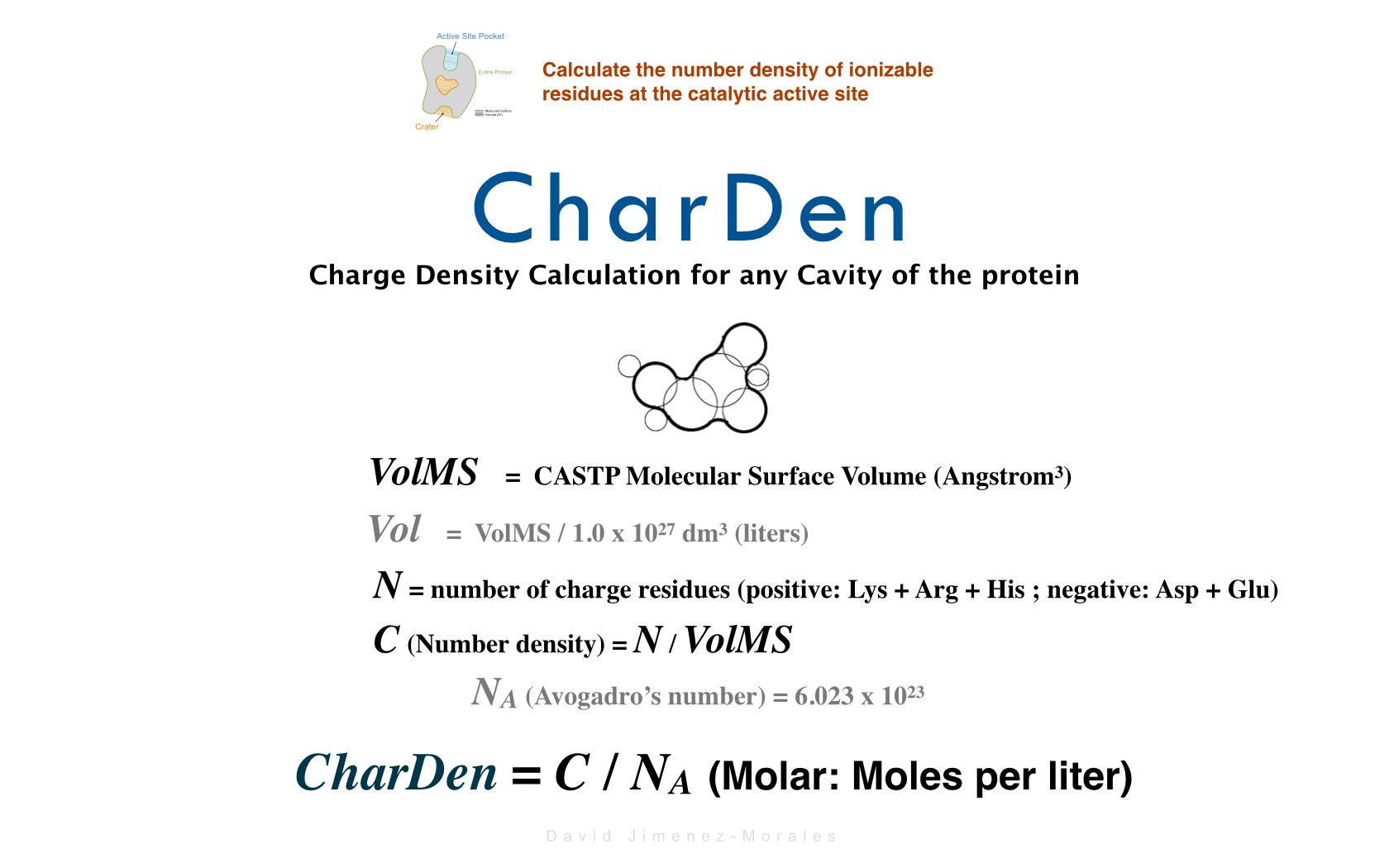
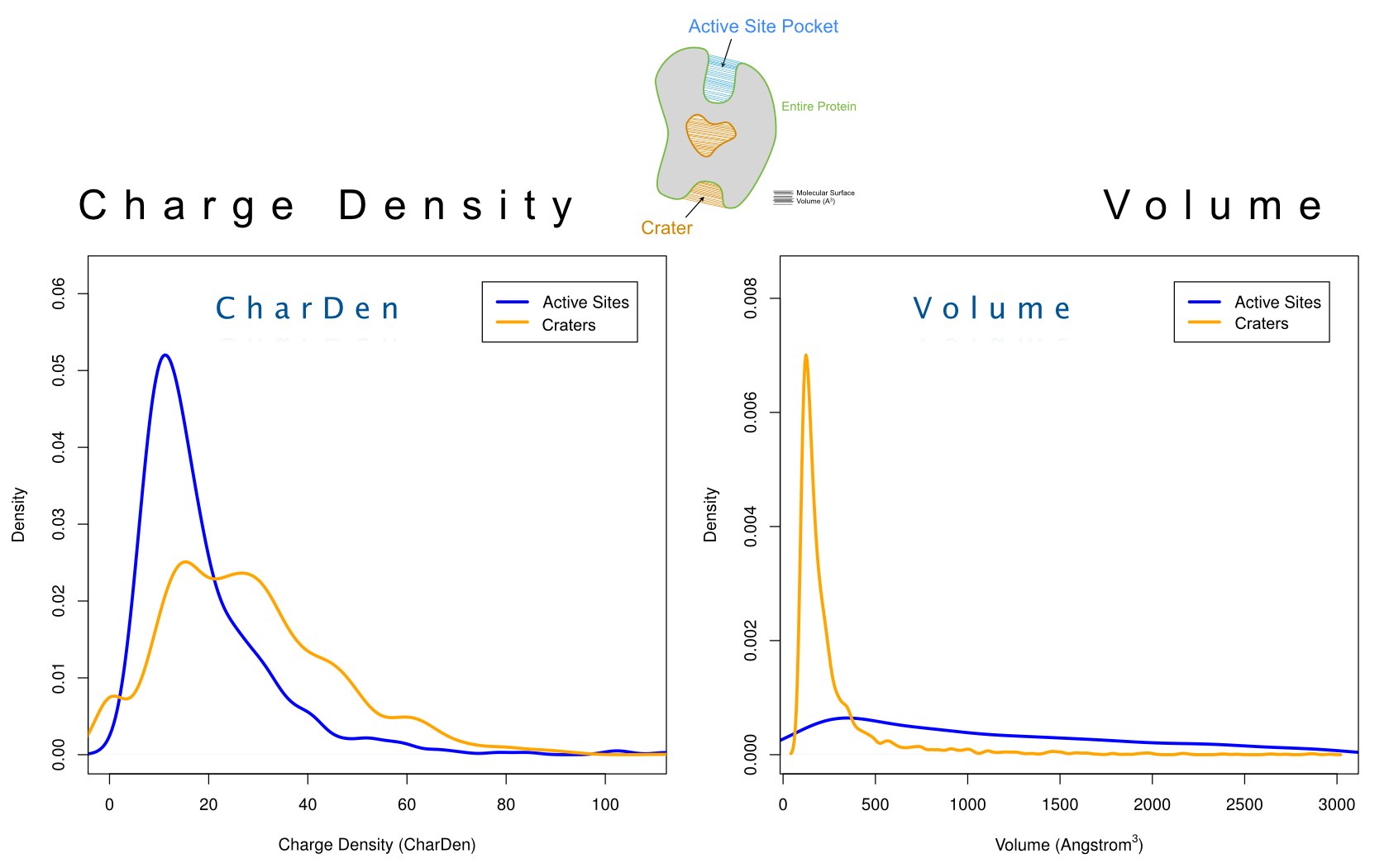
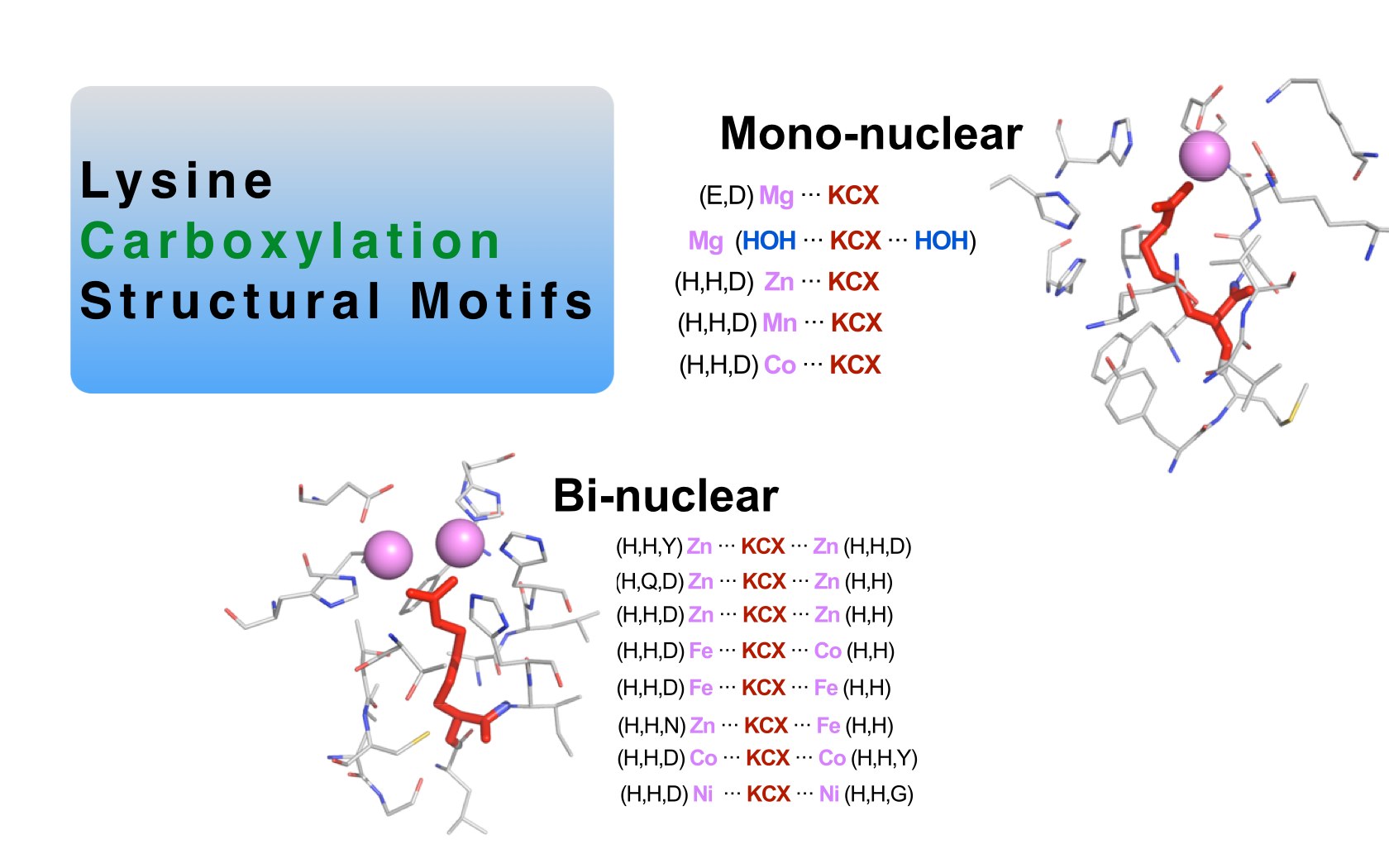
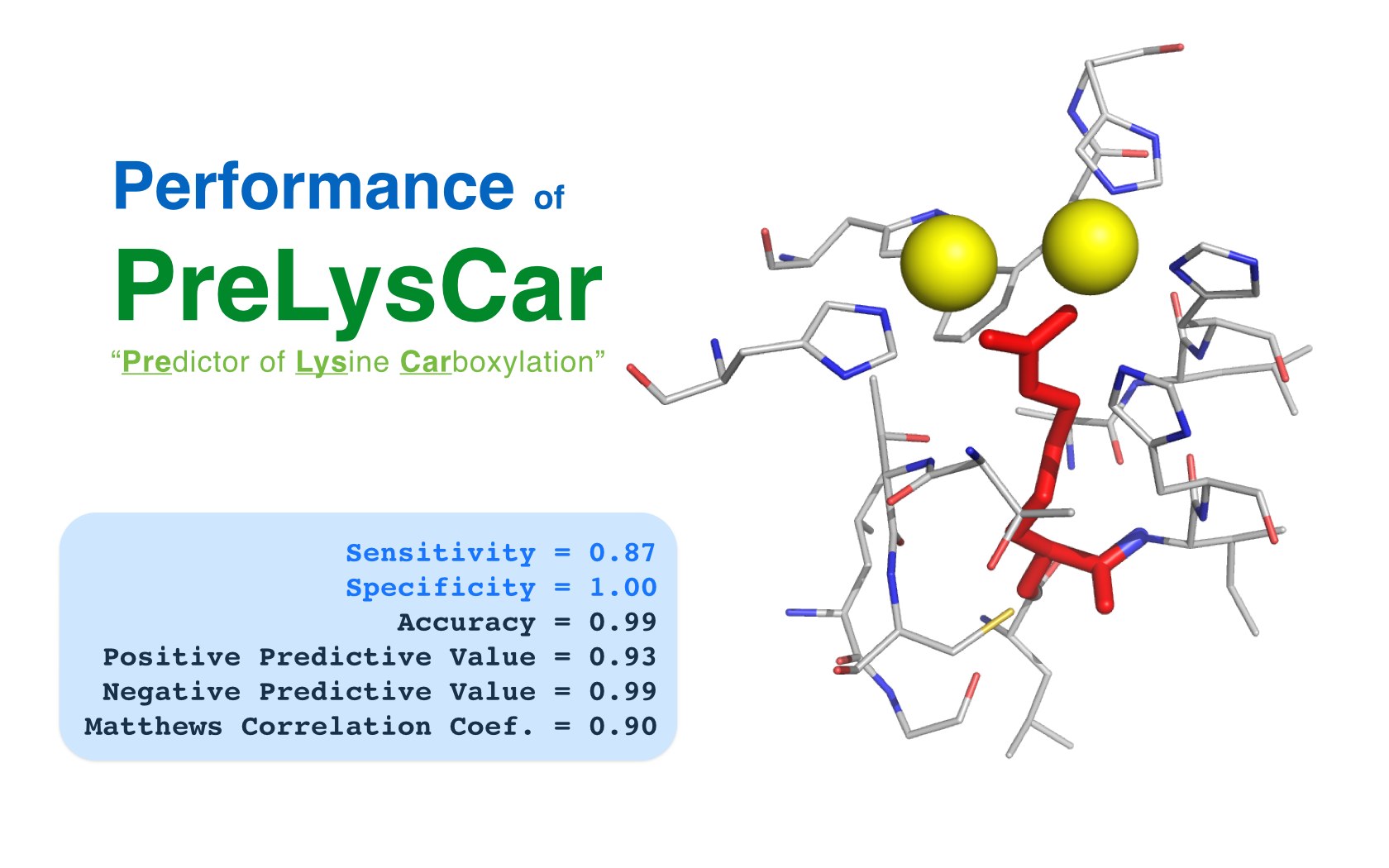
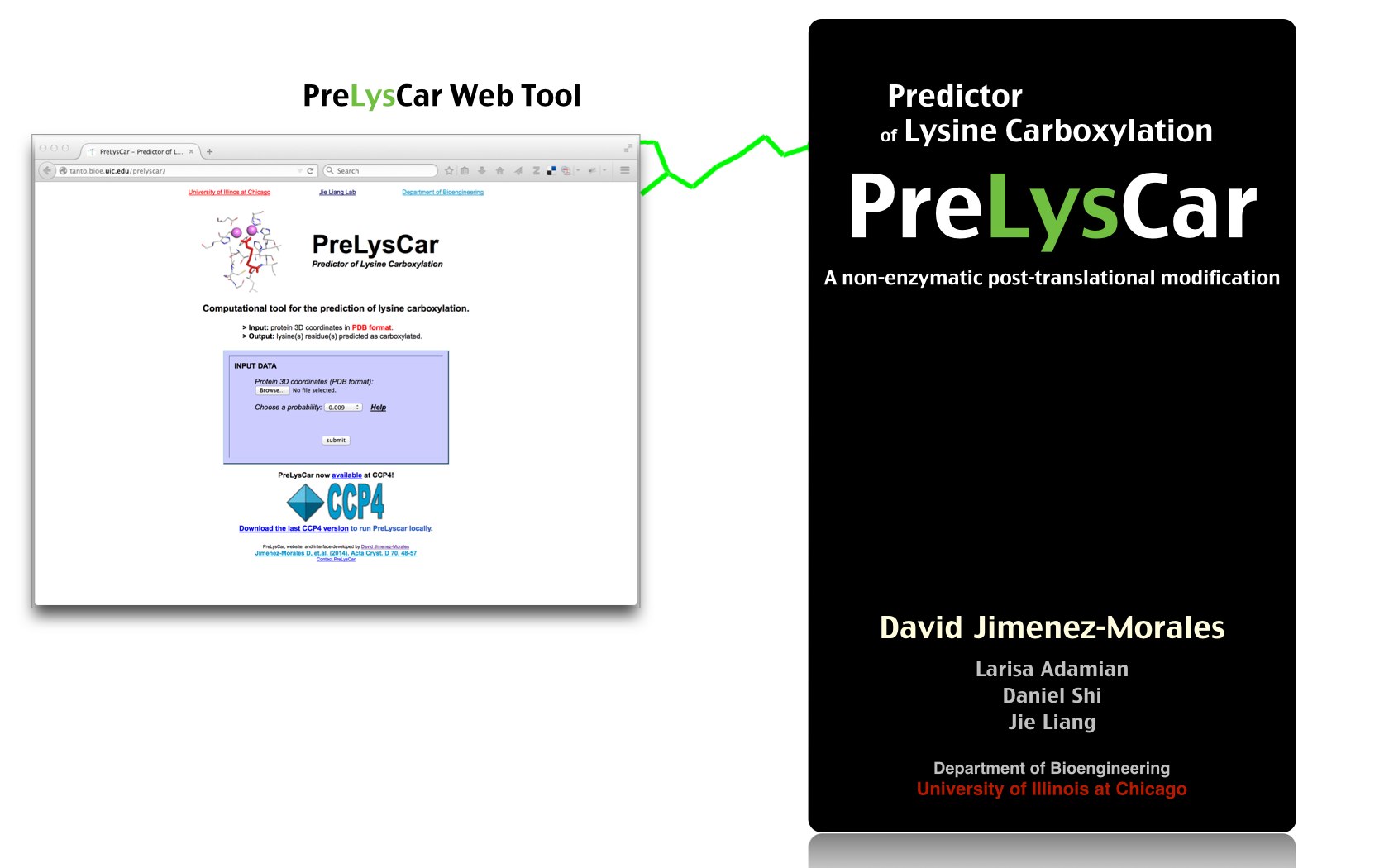
David Jimenez-Morales
Bioinformatics Lead & Research Scientist
Leading computational biology research at Stanford University, specializing in multi-omic data
analysis,
proteomics, and systems biology. PhD in Bioinformatics with extensive experience in developing
bioinformatics tools and pipelines.
But why am
I doing research?